
Figure 20.3.1 Diagram representing an Oral Administration, One Compartment Model
| Boomer Manual and Download | ||||
| PharmPK Listserv and other PK Resources | ||||
| Previous Page | Course Index | Next Page | ||

Figure 20.3.1 Diagram representing an Oral Administration, One Compartment Model
In Figure 20.3.1 only the drug concentrations in the central (plasma or blood) compartment is collected, represented by '*'. An equation which describes this model the bi-exponential equation, Equation 20.3.1

Equation 20.3.1 Drug concentration after Oral Administration
With good data it is possible to estimate ka and kel from the terminal slope and the residual line as described in Chapter 9. The intercept from both these lines provides A.

Equation 20.3.2 Intercept from the Method of Residuals

Figure 20.3.2 Diagram representing and IV Administration, One Compartment Model with Metabolism
Note the samples collected are indicated by the '*'. For example, if only blood was collected but analyzed for drug and metabolite there would be data for these two components of the model. This will limited the number of parameters which can be identified. This can be illustrated by looking at drug concentrations or amounts calculated using this model. Note the data shown in Table 20.3.1 and Table 20.3.2.
| Time (hr) | Cp (mg/L) | Cm (mg/L) | U (mg) |
|---|---|---|---|
| 0 | 10.0 | 0.0 | 0.0 |
| 1 | 7.41 | 0.671 | 8.64 |
| 3 | 4.07 | 0.917 | 19.8 |
| 6 | 1.65 | 0.578 | 27.8 |
| 9 | 0.672 | 0.280 | 31.1 |
| 12 | 0.273 | 0.124 | 32.4 |
Table 20.3.1 Data Calculated with ke = 0.1 hr-1
| Time (hr) | Cp (mg/L) | Cm (mg/L) | U (mg) |
|---|---|---|---|
| 0 | 10.0 | 0.0 | 0.0 |
| 1 | 7.41 | 0.671 | 17.3 |
| 3 | 4.07 | 0.917 | 39.6 |
| 6 | 1.65 | 0.578 | 55.6 |
| 9 | 0.672 | 0.280 | 62.2 |
| 12 | 0.273 | 0.124 | 64.8 |
Table 20.3.2 Data Calculated with ke = 0.2 hr-1
In both tables, above, the data calculated for Cp and Cm are identical even though quite different values of ke were used. The values of km and Vm were different to compensate but we can see that different values of ke could be determined from these data points. The only difference is in the U values. This indicates that a number of parameters including ke, km and Vm may not be identifiable when drug and metabolite concentration in blood or plasma are determined. However, collection of drug amounts in urine has the potential of making these parameters identifiable. This can be further illustrated by plotting these data.

Figure 20.3.3 Linear Plot of Cp and Cm versus Time with different values of ke
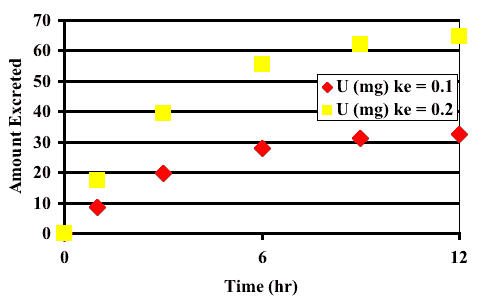
Figure 20.3.4 Linear Plot of U versus Time with different values of ke
Note that in Figure 20.3.3 there is only one line for Cp and Cm, respectively. In Figure 20.3.4 there are two lines for U for the two values of ke. If only Cp and cm are collected ke is non-identifiable and non-observable.

Equation 20.3.3 Rate of Elimination by Michaelis-Menten Kinetics

Figure 20.3.5 Semi-log Plot of Cp versus Time after Three Different Doses
In Figure 20.3.5 three lines are shown after three different doses. This is a semi-log plot. Note the lowest line appears to be a straight line looking like linear kinetics. It would be expected that only a kel value and not Vmax and Km values could be identified from this line of data points. The data collected at higher doses may be more useful in the determination of the Michaelis-Menten parameters. data from all three doses modeled simultaneously would be even better.

Equation 20.3.1 Drug concentration after Oral Administration
If ka is faster than kel then missing early time points can mean that the parameter ka is not locally identifiable. For example, if samples are only collected from four hours on as shown in Figure 20.3.6 ka will be poorly estimated.

Figure 20.3.6 Linear plot of Cp versus Time after Oral Administration
| Time (hr) | Concentration (mg/L) |
|---|---|
| 0 | 0.00 |
| 1 | 5.97 |
| 1.5 | 7.08 |
| 2 | 7.59 |
| 3 | 7.68 |
| 4 | 7.24 |
| 6 | 6.07 |
| 9 | 4.52 |
| 12 | 3.35 |
| 18 | 1.84 |
| 24 | 1.01 |
Table 20.3.3 Data after oral administration - ka > kel
| Time (hr) | Concentration (mg/L) |
|---|---|
| 0 | 0.00 |
| 1 | 5.97 |
| 1.5 | 7.08 |
| 2 | 7.59 |
| 3 | 7.68 |
| 4 | 7.24 |
| 6 | 6.07 |
| 9 | 4.52 |
| 12 | 3.35 |
| 18 | 1.84 |
| 24 | 1.01 |
Table 20.3.4 Data after oral administration - ka < kel
iBook and pdf versions of this material and other PK material is available
Copyright © 2001-2022 David W. A. Bourne (david@boomer.org)